About our study
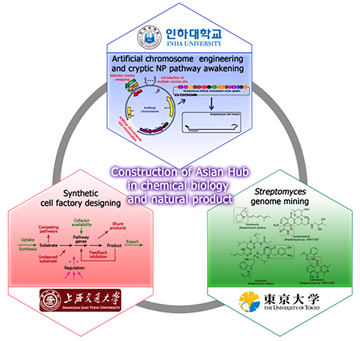
Despite the declining efforts on natural product discovery in the pharmaceutical industry over the past several decades, novel natural product chemical scaffolds have continued to generate human antimicrobial agents, immunomodulators, and antitumor agents, as well as compounds for animal health and plant crop protection. Natural product discovery is currently undergoing a renaissance due to a large extent on the observations from microbial genome sequencing, in which only a fraction of the potential microbial secondary metabolites encoded by actinomycetes and fungi have been discovered. Most secondary metabolite pathways are not expressed under normal laboratory growth conditions. A new discipline, “genome mining”, has emerged and ways to activate the expression of these “cryptic pathways” have been explored.” A recent Special Issue of the J Ind Microbiol Biotechnol (Vol 41, 2014) has covered the recent advances on “Microbial Genome Mining.” In this collaboration project, Japanese side studies biosynthetic enzymes and biosynthesis pathways for several Streptomyces natural products, using some “genome mining” techniques.
Because most microbial biosynthetic genes are clustered within chromosomes, identification of the entire biosynthetic gene cluster is relatively straightforward. Direct cloning combined with heterologous expression of a secondary metabolite biosynthetic gene cluster has become a useful strategy for production improvement and pathway modification of potentially valuable natural products present at minute quantities in original isolates of actinomycetes. However, precise cloning and efficient overexpression of an entire biosynthetic gene cluster remains challenging due to the ineffectiveness of current genetic systems in manipulating large-sized gene clusters for heterologous as well as homologous expression. Meanwhile, most biosynthetic gene clusters are expressed poorly or not at all under laboratory culture conditions. This expression pattern presents a major bottleneck effect for novel natural product discovery, and the awakening of these cryptic pathways is one of the major challenges of genome mining. In this collaboration project, major research objectives in Korean side are to develop a platform technology for the precise cloning and functional overexpression of the entire biosynthetic gene cluster of any potentially-valuable low-titer metabolite in actinomycetes using Streptomyces artificial chromosome system engineering as well as cryptic metabolite pathways awakening techniques.
Since 1980’s, hundreds of non-natural natural products have been obtained through combinatorial biosynthesis, which provides a powerful and promising alternative to the traditional screening of drug leads. However, in most cases the yields of engineered compounds are too low for further characterization and clinical trials due to following possible reasons. Firstly, most of the wild-type antibiotic producers have relatively low productivity and not been fully optimized for the production. Secondly, the introduced genes are not highly expressed due to the lack of efficient promoters and regulatory elements. Thirdly, downstream processing enzymes have low catalytic efficiency toward the substrates with altered structures. Therefore, the advent of synthetic biology will shed new lights on alleviating these hurdles and significantly increasing the yields of engineered derivatives. In order to provide a suitable background for antibiotic high yield and instead of performing multiple engineering of the wild-type producers, other hosts derived from industrial high-yielding strains producing similar compounds become ideal chassis for heterologous expression of the interested gene clusters. The high-yielding mechanisms for aminocyclitol validamycin, polyether salinomycin, polyene pimaricin, non-ribosomal peptide vancomycin have been studied. These strains are superior to the wild-type and most other antibiotic producers with excellent fermentation features, high gene expressions levels, abundant precursors and cofactors for corresponding class of natural products. Therefore, the main assignment of Chinese side is to systematically engineer these strains into convenient chassis.